Code
#install uunet
import uunet.multinet as ml
n = ml.read('tutorial/example1.txt')
ml.plot(n, vertex_labels_bbox = {"boxstyle":'round4', "fc":'white'})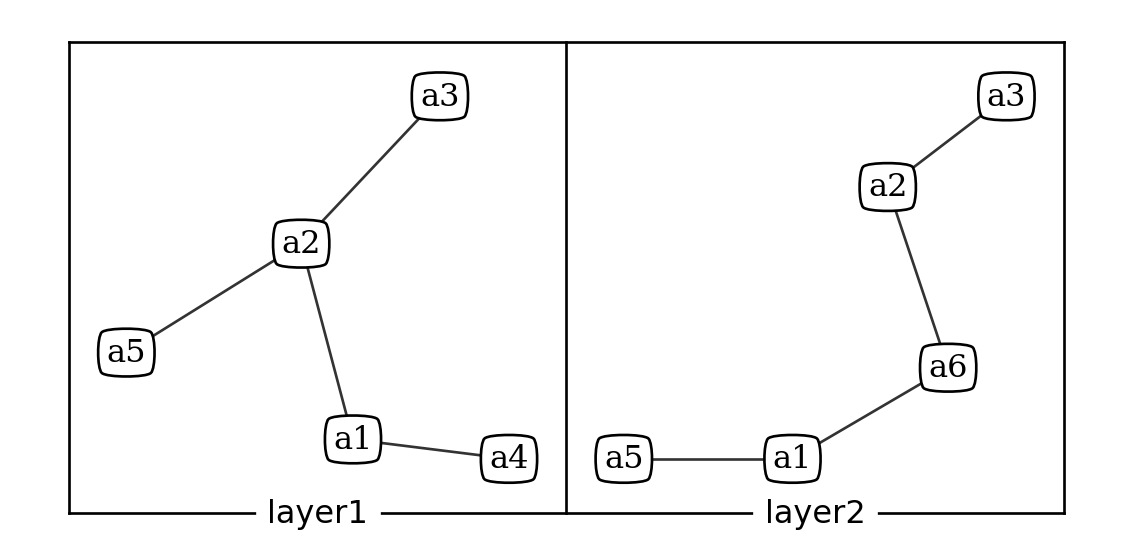
Harun Pirim
October 17, 2023
First introduced under social science domain, multilayer networks represented different type of relationships of the same nodes. The representation is now generalized to allow diffent nodes at each layer, different relathionships and connections between layers. Transportation networks, social networks, biological networks, and financial networks are examples of multilayer networks.
I will be using the convention adopted in the first reference. We can define multilayer network
\(M = (Y, \vec{G}, G)\)
where \(Y\) is the set of layers, is the ordered list of networks, and \(G\) is the list of bipartite networks that represent relationships between pairs of layers.
Multiplex networks are multilayer networks where the set nodes is the same for all layers. They are also called replica nodes if interlinks are allowed between layers. Without interlinks a multiplex can be represented as
\(M = (Y, \vec{G})\)
where \(Y\) is the set of layers and is the ordered list of networks. Each network contains the same set of nodes.
One way of analyzing multiplex networks is flattening layers and analyzing the resulting single layer network. This approach is called single layer projection. Edge weights can be added to reflect the number of layers that contain the edge.
Degree standard deviation of an actor through layers is defined as
\(\sigma_{k} = \sqrt{\frac{1}{L-1}\sum_{\alpha=1}^{L}(k_{\alpha}-\bar{k})^2}\)
where \(k_{\alpha}\) is the degree of the actor in layer \(\alpha\), \(\bar{k}\) is the average degree of the actor, and \(L\) is the number of layers.
uunet Python package can be used to analyze multiplex networks. Detailed documentation and a tutorial can be found at uunet

Helper function defined to convert the output of the library to pandas dataframes.
Use networkx objects as layers of the multiplex network
| from_actor | from_layer | to_actor | to_layer | dir | |
|---|---|---|---|---|---|
| 0 | A | layer1 | C | layer1 | False |
| 1 | A | layer1 | B | layer1 | False |
| 2 | B | layer1 | C | layer1 | False |
| 3 | A | layer2 | C | layer2 | False |
The library provides a multiplex mixture growth model based on some evolution functions. It’s limited to ER and PA models for now. A layer can grow internally or externally based on probabilities. If externally effected from other layers, dependency matrix can be used to define the dependency between layers.
models_mix = [ ml.evolution_pa(3, 1), ml.evolution_er(100), ml.evolution_er(100) ]
pr_internal = [1, .2, 1]
pr_external = [0, .8, 0]
dependency = [ [1, 0, 0], [0.5, 0, 0.5], [0, 0, 1] ]
generated_mix = ml.grow(100, 150, models_mix, pr_internal, pr_external, dependency)
l = ml.layout_multiforce(generated_mix, gravity = [.5])
ver = ml.vertices(generated_mix)
deg = [ml.degree(generated_mix, [a], [l])[0] for a,l in zip(ver['actor'], ver['layer'])]
ml.plot(generated_mix, layout = l, vertex_labels=[], vertex_size=deg)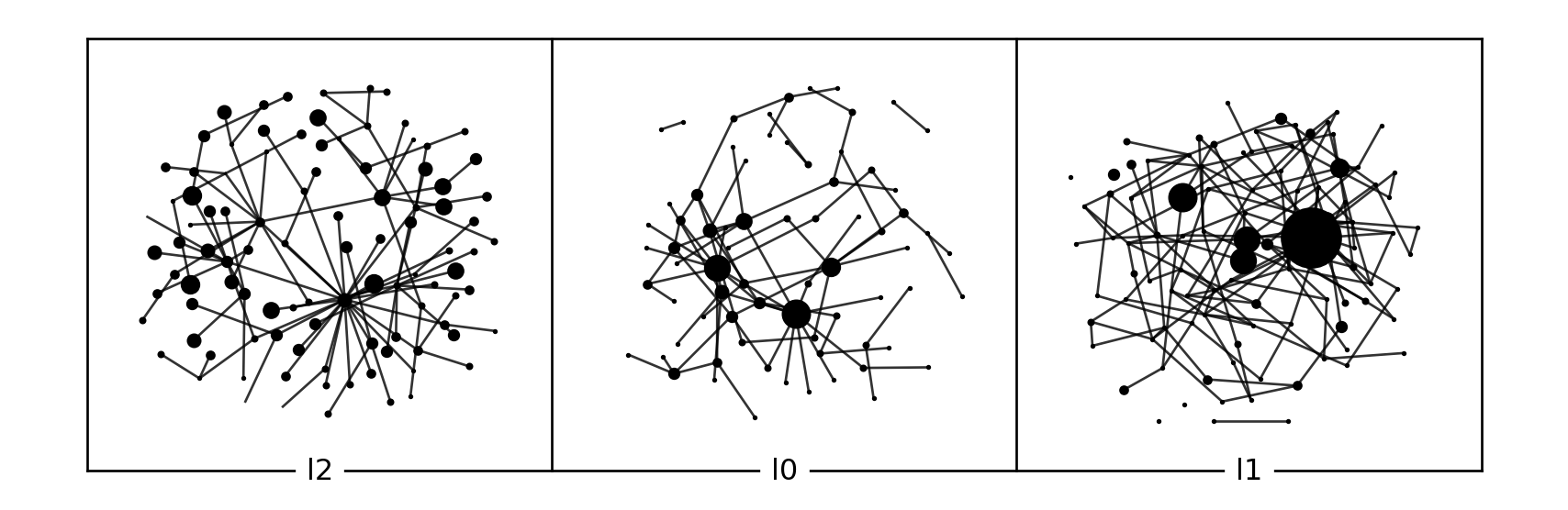
It is also possible to read existing datasets in the library.
You can merge some layers into a single layer.
['lunch', 'facebook', 'coauthor', 'leisure', 'work', 'offline']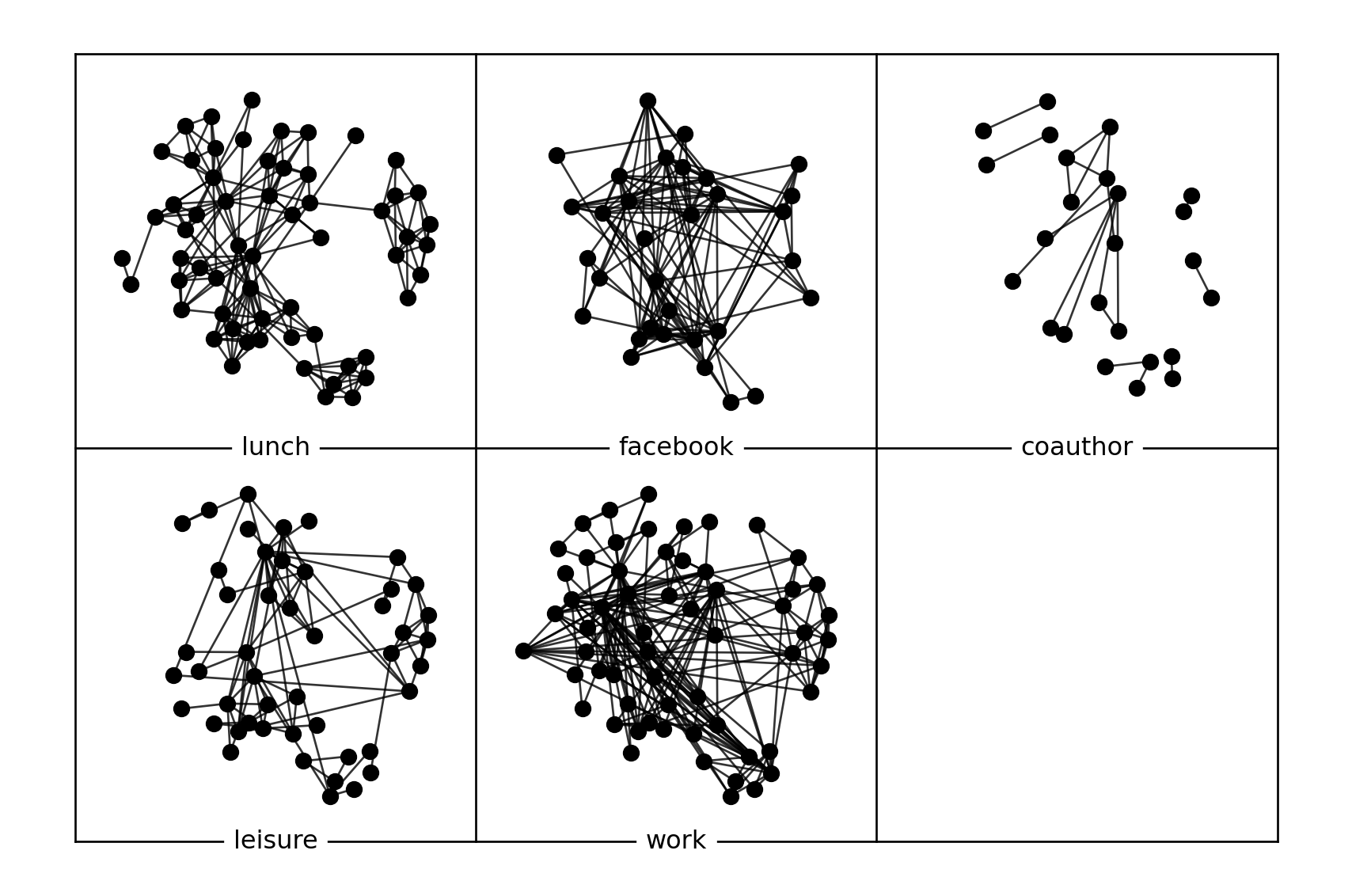
Some summary network metrics can be obtained from the library. \(n,m\) are number of nodes and edges, \(nc\) is number of components, \(slc\) is the size of larges component, \(dens\) is the density, \(cc\) is the average clustering coefficient, \(apl\) is the average shortest path length, \(dia\) is the diameter of the layer.
| layer | n | m | dir | nc | slc | dens | cc | apl | dia | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | lunch | 60 | 193 | False | 1 | 60 | 0.109040 | 0.568926 | 3.188701 | 7 |
| 1 | 32 | 124 | False | 1 | 32 | 0.250000 | 0.480569 | 1.955645 | 4 | |
| 2 | coauthor | 25 | 21 | False | 8 | 6 | 0.070000 | 0.428571 | 1.666667 | 3 |
| 3 | leisure | 47 | 88 | False | 2 | 44 | 0.081406 | 0.343066 | 3.122622 | 8 |
| 4 | work | 60 | 194 | False | 1 | 60 | 0.109605 | 0.338786 | 2.390395 | 4 |
Other network statistics can be obtained converting multiplex object to networkx object.
0.0027009183122244525Layers can be compared in pairs based on difference/similarity of node/edge properties. The formulations are explained in the reference 3.
| lunch | coauthor | leisure | work | ||
|---|---|---|---|---|---|
| lunch | 0.000000 | 0.420768 | 2.896653 | 1.328825 | 0.837241 |
| 0.420768 | 0.000000 | 2.021401 | 1.017798 | 0.710679 | |
| coauthor | 2.896653 | 2.021401 | 0.000000 | 0.452108 | 0.591749 |
| leisure | 1.328825 | 1.017798 | 0.452108 | 0.000000 | 0.211845 |
| work | 0.837241 | 0.710679 | 0.591749 | 0.211845 | 0.000000 |
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | 1.000000 | 0.533333 | 0.416667 | 0.783333 | 0.967213 |
| 1 | 0.533333 | 1.000000 | 0.295455 | 0.519231 | 0.533333 |
| 2 | 0.416667 | 0.295455 | 1.000000 | 0.411765 | 0.416667 |
| 3 | 0.783333 | 0.519231 | 0.411765 | 1.000000 | 0.783333 |
| 4 | 0.967213 | 0.533333 | 0.416667 | 0.783333 | 1.000000 |
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | 1.000000 | 0.312560 | 0.148637 | 0.281517 | 0.246475 |
| 1 | 0.312560 | 1.000000 | 0.547277 | 0.378174 | 0.540601 |
| 2 | 0.148637 | 0.547277 | 1.000000 | 0.480845 | 0.427194 |
| 3 | 0.281517 | 0.378174 | 0.480845 | 1.000000 | 0.068050 |
| 4 | 0.246475 | 0.540601 | 0.427194 | 0.068050 | 1.000000 |
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | 1.000000 | 0.178439 | 0.064677 | 0.277273 | 0.339100 |
| 1 | 0.178439 | 1.000000 | 0.058394 | 0.158470 | 0.186567 |
| 2 | 0.064677 | 0.058394 | 1.000000 | 0.101010 | 0.091371 |
| 3 | 0.277273 | 0.158470 | 0.101010 | 1.000000 | 0.205128 |
| 4 | 0.339100 | 0.186567 | 0.091371 | 0.205128 | 1.000000 |
Degrees and degree deviations of actors can be obtained.
[[49, 'U4'], [47, 'U67'], [46, 'U91'], [44, 'U79'], [44, 'U123']]| actor | lunch | coauthor | leisure | work | ||
|---|---|---|---|---|---|---|
| 0 | U4 | 15 | 12 | NaN | 1.0 | 21 |
| 1 | U67 | 12 | 13 | NaN | 2.0 | 20 |
| 2 | U91 | 7 | 14 | 3.0 | 14.0 | 8 |
| 3 | U79 | 13 | 15 | NaN | 7.0 | 9 |
| 4 | U123 | 6 | 11 | NaN | NaN | 27 |
| actor | dd | |
|---|---|---|
| 0 | U4 | 8.133880 |
| 1 | U67 | 7.418895 |
| 2 | U91 | 4.261455 |
| 3 | U79 | 5.230679 |
| 4 | U123 | 9.987993 |
Neigborhood centralities can be calculated. See the difference from degrees.
[22]Neigborhood centrality exclusive to the queried layers can be calculated.
Relevance is the ratio between neighborhood centrality of the queried layers and all layers.
| actor | lunch | coauthor | leisure | work | ||
|---|---|---|---|---|---|---|
| 0 | U4 | 0.576923 | 0.461538 | NaN | 0.038462 | 0.807692 |
| 1 | U67 | 0.521739 | 0.565217 | NaN | 0.086957 | 0.869565 |
| 2 | U91 | 0.318182 | 0.636364 | 0.136364 | 0.636364 | 0.363636 |
| 3 | U79 | 0.541667 | 0.625000 | NaN | 0.291667 | 0.375000 |
| 4 | U123 | 0.206897 | 0.379310 | NaN | NaN | 0.931034 |
Multiplex networks are special cases of multilayer networks. Multilayer networks can be defined with different nodes at each layer. Pymnet libary can be used to visulaize and analyze multilayer networks.
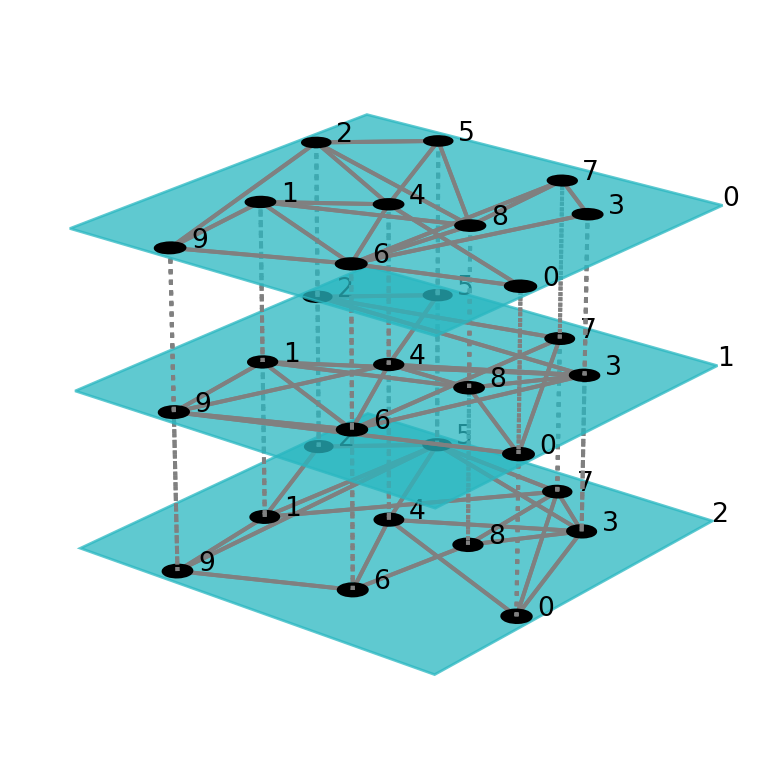
gml = pn.er(10,3*[0.4])
#list(gml)
#list(gml.edges)
#list(gml.iter_layers())
#list(gml.iter_node_layers())
#list(gml[0,0])
#gml[0,0].deg()
#gml[0,0].str()
#pn.degs(gml)
#pn.density(gml)
#pn.cc_barrat(gml,(0,0))
#pn.multiplex_density(gml)
#pn.supra_adjacency_matrix(gml)
#help(pn.models)
#help(pn.netio)1: Bianconi, Ginestra. Multilayer Networks. Available from: VitalSource Bookshelf, Oxford University Press Academic UK, 2018.
3: Bródka, Piotr, et al. “Quantifying layer similarity in multiplex networks: a systematic study.” Royal Society open science 5.8 (2018): 171747.